A deep understanding of the relation between genetic mutations and immune system response of the attacked organism is vital for the development of effective vaccinations for specific variations of the influenza virus.
The formulation of a theory that associates immunological response to phylogenetic evolution of the virus stands on the definition of a genetic distance – calculated between aminoacid sequences – able to quantify antigenic diversity among the corresponding pathogens.
Despite the wide scientific success that present models obtains in studying the dynamic of viral infections, in many cases the methodology stands on genetic distances that only count the number of mutations, neglecting their position in the aminoacids chain. Several ideas, on the other side, aim at recognizing the modular schemes in the mutations and expressing the genetic distance based on the semantic variation brought at a global level by the single point mutation.
An example will clarify the concept. Let us imagine talking about words instead of aminoacids sequences, and about meaning instead of viral pathogens. Let us consider, for example, the words “cane” and “pane”. According to the classic genetic distance, these two words have distance 1, since there is only one mutation that distinguishes them. Now, let us imagine causally changing the order of the letters in the words, in the same way in both, obtaining the two new words “aenc” and “aenp”. The classic genetic distance is still 1, since this distance does not change if the order is modified and it is therefore unable to acknowledge that the two words are now meaningless.
It is important to formulate a genetic distance able to detect meaning variations (i.e. in the virus structure) starting from the variations in the letter sequence (i.e. aminoacids).
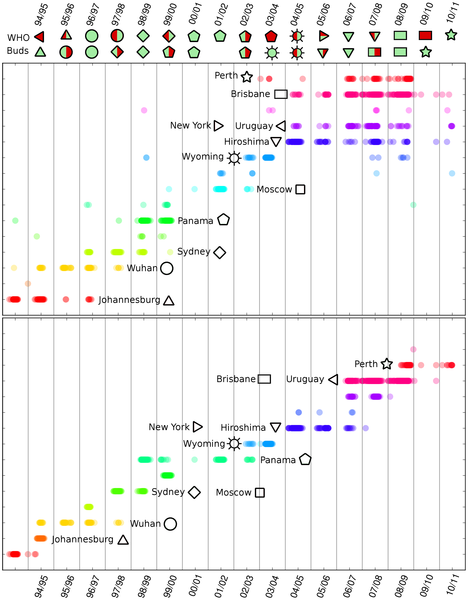
The chart shows haemoagglutinin (HA) genetic evolution of the influenza virus belonging to H3N2 family (click on the chart to show the detailed study).
Haemoagglutinin is an antigen glycoprotein present on the virus surface and it is responsible, together with neuraminidase, of virus adhesion to cells. For this reason, it is recognized by the immune system and it is the most subjected to genetic drift. This analysis is based on the theory developed by Riccardo Scalco, Mario Casartelli and Raffaella Burioni, University of Parma.
Different aminoacids sequences, from GISAID database, have been compared using a genetic distance defined by Shannon mutual conditional entropy between partitions of abstract probability spaces. It is assumed that this distance can observe biologically relevant changes in the amino acid redisposition proposed by the virus evolution. The evolution emerging by the proposed analysis showed already to be in optimal accordance with the epidemiological history of the viral pathogen.
The chart’s interpretation is the following. Every HA aminoacid sequence, associated to an isolated virus (i.e. a virus extracted by a patient’s blood) is identified as a coloured dot. The position on the abscissa axis indicates the isolation date, whilst the position on the ordinate axis allows grouping together isolated viruses belonging to the same subfamily (according to the used genetic distance). In other words, isolated viruses belonging to different subfamilies have different positions on the ordinate axis. The colours have the only visual function to allow easier recognition of the subfamilies.
It is then possible to make some important observations. First, every winter several subfamilies are present: one is usually predominant, while either the others are subfamilies already appeared in the previous years – and gradually disappearing – or they are subfamilies never appeared before. The analysis also suggests searching for the predominant virus of the following season within the new subfamilies. It is in fact easy to observe that the principal subfamily of a certain season has often precursors in the previous seasons.
- Burioni R, Scalco R, Casartelli M. Rohlin Distance and the Evolution of Influenza A Virus: Weak Attractors and Precursors. PlosOne 2011. DOI: 10.1371/journal.pone.0027924
- Pybus O, Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Nat Rev Genet 2009; 10: 540–550.
- Recker M, Pybus O, Nee S, Gupta S. The generation of inuenza outbreaks by a network of host immune responses against a limited set of antigenic types. Proc Natl Acad Sci USA 2007; 104: 7711–7716.
- Hamming R. Error detecting and error correcting codes. Bell System Technical Journal 1950; 29: 147–160.
- Khinchin A. Mathematical Foundations of Information Theory. Dover, New York, 1957.
- Study website